Illumina® Website (March 2018):
"Index hopping or index switching is a known phenomenon that has impacted NGS technologies from the time sample multiplexing was developed. It causes a specific type of misassignment that results in the incorrect assignment of libraries from the expected index to a different index (in the multiplexed pool).
Index hopping can be seen at slightly elevated levels on instruments using patterned flow cells with exclusion amplification chemistry versus those that do not use patterned flow cells. Libraries with higher levels of free adapters will see higher levels of index hopping."
Index hopping can be seen at slightly elevated levels on instruments using patterned flow cells with exclusion amplification chemistry versus those that do not use patterned flow cells. Libraries with higher levels of free adapters will see higher levels of index hopping."
The phenomenon has been described in a preprint paper (by Rahul Sinha et al.) that suggests the issue might occur during the act of sequencing itself, with free oligos in the pool somehow associating with the fragments being sequenced and causing the fragment to be identified with the index of the free oligo as opposed to it’s true original sequence.
Any single index system may be subject to this phenomenon, but the NEXTFLEX® unique dual index barcodes have been designed to specifically mitigate the index hopping phenomenon. With unique dual index barcodes, if a free oligo associates with a fragment causing a "change" in the index of the read, the other index associated with the fragment will show the read is incorrect and prevent it from being associated with a given sample.
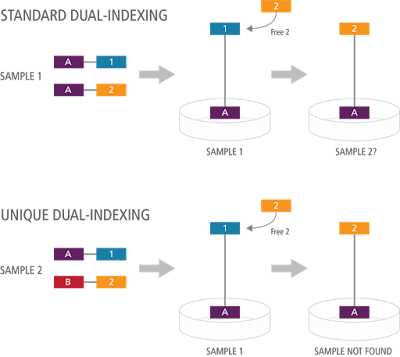
It is important to note however that not all dual index systems would address this problem, as standard dual indexes using a combinatorial approach (using the same first index across sets of a second index) would still lead to the incorrect allocation of some reads as shown below.
In the standard dual indexing example below (combinatorial dual indexing), the fragment was originally indexed with the blue and purple indexes, but it now being identified as indexed with the gold and purple indexes. However because there is also a sample present using the gold and purple index combination, this read is now going to be incorrectly included in that other sample.
However if this same situation occurs with the UNIQUE dual indexes, the resultant contaminant read will not be added to another sample present. The incorrect read will still occur, but this time it isn’t reassigned to another sample present, as no sample contains that combination of indexes. Both indexes are unique so the issue is avoided.
Index mis-assignment can lead to increased false positive rates, which are especially detrimental to sensitive applications.
Multiplexing with NEXTFLEX® unique dual index barcodes significantly increases processing capacity while reducing costs by allowing the user to pool multiple libraries in a single flow cell lane, whilst providing unprecedented data security in sequencing applications.
Written by Rick Bhatt
If you like my post why not connect to me on LinkedIn.